
Graph distance index

Descriptor Category : topological
c++ entity: functor
Description
The graph distance count of nth order nf is defined as the total number of
distances equal to n in the graph.
The graph distance code, is the ordered sequence of graph distance counts
<1f, 2f, 3f,..., df>, where d is the topological diameter.
The graph distance index is defined as the squared sum of all graph distance counts:
Equation 1: Graph distance index.
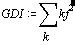 |
Prototype
template <class Molecule>
struct graph_distance_index : std::unary_function<Molecule,size_t> {
result_type operator()(typename boost::call_traits<argument_type>::param_type m);
};
Where defined
morpho/cdl/descriptors/topological.hpp
Namespace
morpho::cdl
Inherits from
std::unary_function<Molecule,size_t>
Arguments
| Argument | Model of |
| Molecule |
cdl::molecule |
Example
// suppose you're streaming a mol format from the stdin:
typedef desc_molecular_properties<> desc_mol_props_t;
typedef boost::property<mol_propsS, molecular_properties<>,
boost::property<descriptors_mol_propsS, desc_mol_props_t> > descr_props_t;
typedef molecule<double,descr_props_t> M;
morpho::cdl::nail_juice<M> j;
morpho::cdl::get_juice_from_stream(std::cin, j, 0, sdf_formatT());
M mol(j);
morpho::cdl::graph_distance_index<M> GDI;
std::cout << "Graph distance index of my mol: ";
std::cout << GDI(mol) << '\n';
Related Items
topological radius and diameter
polarity number
References
- Todeschini, R.; Consonni, V. "Handbook of Molecular Descriptors". Wiley-VCH,
Methods and Principles in Medicinal Chemistry. Volume 11. 2000.
Copyright (c) Vladimir Josef Sykora and Morphochem AG 2003

